Through gene regulation, controlling which genes are active and which proteins are produced at any given time is crucial for cells to function properly. Improper regulation can lead to serious disease states, including developmental disorders and diseases such as cancer, when cells grow uncontrollably or fail to respond properly to signals. Efficient gene regulation enables organisms to efficiently manage growth, repair, and immune response. In this context, it is essential to gain a better understanding of how this complex regulation works, which was the focus of a PRX Life study published in late August 2024.
Our study is the first in which anyone has explored the idea that insulators may interact weakly with surrounding chromatin rather than with themselves.
The study, led by Umeå researchers Lucas Hedström and Ludvig Lissana in collaboration with Ralf Metzler at the University of Potsdam, provides a new perspective on how cells control gene expression using spatially separated enhancers and insulators. Enhancers are short DNA sequences that enhance gene activity by binding to proteins at promoter sites, while insulators act as physical barriers that prevent enhancers from doing so. Conventional models suggest that insulators interact directly with each other to reduce the connections between enhancers and promoters. However, the study authors now challenge this idea by introducing a stochastic model in which insulators bind weakly to the surrounding DNA rather than to each other.
In simpler terms, says Ludwig Liszana, associate professor in the Department of Physics and IceLab, you can imagine two pieces of tape.
– The tape does not need to stick to another tape – The tape can stick weakly to other surfaces as well. By insulators we mean that they do not need to bind to each other – contact with chromatin, the DNA strand, is enough to prevent the enhancer from promoting expression of the gene as long as it actually blocks it.
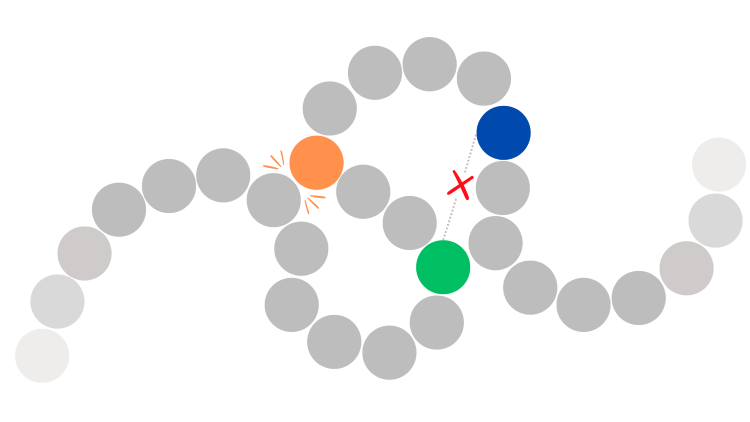
Illustration of DNA where the orange insulator sequence creates a physical block by binding to the DNA strand and preventing the green enhancer sequence from binding to the blue promoter sequence to enhance gene expression.
Lucas Hedström, a PhD student in the Department of Physics and author of the study, explains how they came up with this model:
– Yuri Schwartz's group recently presented new experimental data, published in Science Advances in February 2023, that challenged the current model of how insulators work. With this in mind, we built a new model to show that perhaps not all cell regulatory processes work in the same way, but rather that multiple mechanisms may be at play.
By simulating these regulatory interactions, the researchers found good agreement with experimental data on the Hi-C interaction from screening hundreds of insulating elements in fruit flies from Yuri Schwartz’s study. The researchers also found significant variation in how long it took for enhancers to contact their target genes, suggesting that gene regulation occurs on a more unpredictable time scale than previously thought. This variation could have important implications for understanding the dynamics of gene expression in complex organisms.
Study author Ludwig Liszana explains:
“Our study is the first to explore the idea that insulators may interact weakly with surrounding chromatin rather than with themselves. Our work provides new insights into the relationship between local 3D folding of the genome and gene activation, which could reshape the way we approach gene regulation research. These findings could also lead to breakthroughs in how we understand the regulation of developmentally relevant gene clusters that require precise control of gene expression, and potentially in gene therapy and synthetic biology.”





More Stories
The contribution of virtual reality to research in medicine and health
The sun could hit the Internet on Earth
In memory of Jens Jørgen Jørgensen